Rockfish DNA Reveals Genetic Clues For Longevity
A study in Science identifies genes related to immunity, DNA repair, and nutrient sensing as genetic drivers for rockfish lifespans exceeding 200 years.
Highlights
· Kolora and colleagues sequenced over 100 fish from 88 rockfish species to identify genes that evolved and drive lifespan.
· Genes linked to a longer lifespan were related to immunity and inflammation, size and environmental adaptations, and DNA repair pathways.
· Duplications in the immune modulatory butyrophilin gene family were found in long-lived species.
The lifespan of animals has a vast range, varying from days and weeks to hundreds of years. A fish called a pygmy goby has a 5-week life cycle, whereas some Greenland sharks have been estimated to live for about 400 years. Scientists are looking for the DNA changes that occurred during the evolution of these creatures to create such diversity in lifespan to understand the genetic drivers of longevity better. In other words, we’re on the hunt for the genes and modifications to their sequences that may be at the root of living for centuries.
In an article published in Science, researchers studying over a hundred highly related rockfish that come in many different sizes, live at different depths, and have a huge range of lifespans pin down several genetic drivers of longevity. Kolora and colleagues from the University of California, Berkeley, reveal selective DNA signatures in pathways that underlie several “hallmarks of aging,” providing clues as to what genes affect our longevity. Many of the identified pathways are conserved across all animals, such as DNA damage and nutrient-sensing pathways, as well as in vertebrate-specific hallmarks like immunity and inflammation.
“There is an opportunity here to look in nature and see how natural adaptations have shaped lifespan and to think about how those same sorts of genes are acting in our own bodies,” said senior author Peter Sudmant, an assistant professor at UC Berkeley.
Watching Evolution at Hyperspeed
Rockfishes (genus Sebastes) of the Pacific Ocean exhibit huge lifespan ranges. Highly related rockfish species live anywhere from 11 years (Sebastes minor) to greater than 200 years (rougheye rockfish, Sebastes aleutianus). More than 120 different species of rockfish are found throughout the northeast and the northwest Pacific Ocean. This abundance of species with vastly differing life histories represents an example of repeated, recent adaptations that have reshaped longevity characteristics.
“You could think of rockfish as sort of the perfect storm. in some ways, both on an individual level — having individual fish able to live for a really long time because of size and depth adaptations — but also just having all these different species that are showing these different trends,” said Sudmant. “They’re a perfect set of individuals to look at, where other people just had a single species to look at.”
To dissect the genetic underpinnings of life-span variation and adaptation, Kolora and colleagues sequenced and assembled the genomes of 102 rockfish individuals encompassing 88 different species from scratch — that is, these genomes had never been sequenced before. The UC Berkeley researchers also constructed a highly detailed evolutionary map of the rockfish species. With this tree of rockfish evolution with different lifespans, the researchers created a foundation for determining the genetic underpinnings of lifespan in Pacific rockfishes.
“In these rockfish, we can actually watch this evolution happening over this 10-million-year time period, and we observe that when some species evolve a short lifespan, their population sizes expand, and when they evolve a long lifespan, their population sizes contract,” he said. “We can see a signature of that in their genomes, in the genetic variation that exists in these species. So, there is a consequence to adapting to long and short life.”
Genetic underpinnings of life-span adaptations
Kolora and colleagues found gene signatures that were linked to the evolution of the fish. For example, whereas no pathways were enriched in positively selected genes — genes that are kept in evolution — in short-lived species, positively selected genes in long-lived species were enriched for DNA double-stranded break repair pathways. They also discovered 91 genes significantly associated with life span, including candidates with roles in cell growth and proliferation, DNA repair, and suppression of programmed cell death.
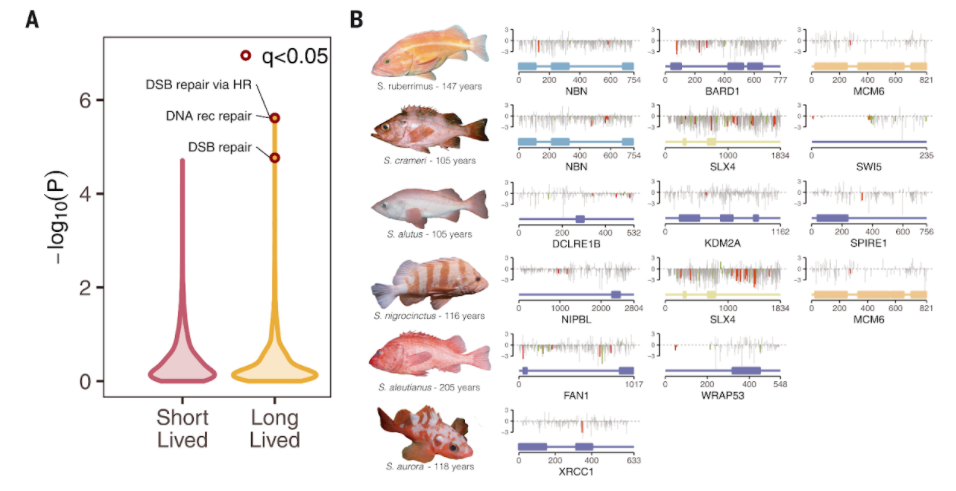
Kolora and colleagues also found that there’s a major link between lifespan and the size and environment of the fish as well as genes involved in those processes. Since rockfish lifespan is correlated with body size and environmental factors, such as depth, some of these genes linked to lifespan may act by influencing growth and size or may facilitate adaptations to environments that promote longevity.
“We can explain 60% of the variation in lifespan just by looking at the size at maturity and the depth at which a fish lives,” said Sudmant. “So, you can predict lifespan with pretty high accuracy just from these factors. This allowed us to identify the genes that allow them to do those things.”
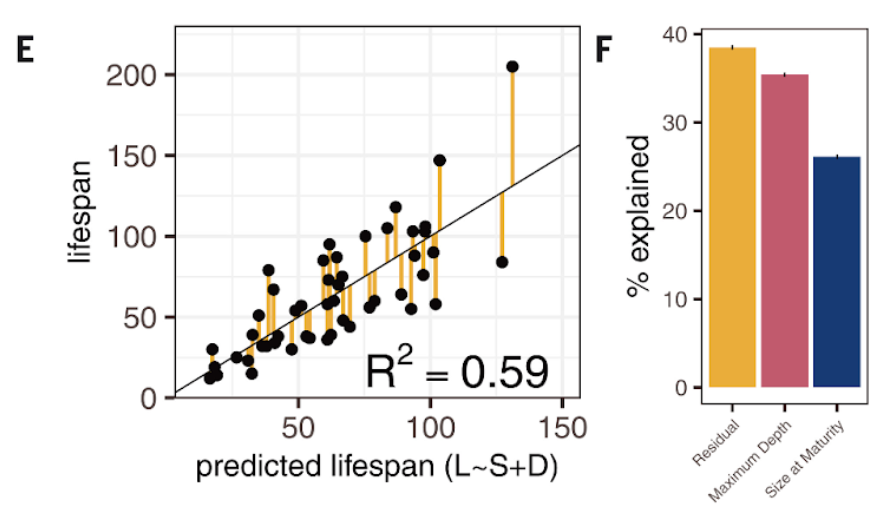
Independent of size at maturity or depth, Kolora and colleagues identified 56 genes associated with lifespan, such as those related to nutrient sensing and insulin/glucose signaling. The rate of evolution for most of these genes was negatively correlated with life span, emphasizing the importance of nutrient-sensing maintenance in long-lived species. They also identified several genes that have roles in life-span extension across many organisms, such as ones associated with reproductive aging in mice, antiviral innate immunity, and tumor suppression.
“In this study, we identified both the genetic causes and consequences of adaptation to extreme lifespan,” said Sudmant, UC Berkeley assistant professor of integrative biology. “It’s very exciting to be able to look at a group of species and see how their [lifespan] has been shaped through time and the genetic changes that drive that [trait], and simultaneously, how that [trait] then feeds back and influences the genetic diversity of that population.”
These analyses highlight the genetic innovations that underlie life-history trait adaptations and, in turn, how they shape genomic diversity.